Laboratory of Photosynthesis
Josef Komenda`s group
Biogenesis of Photosystem II
Research Projects
Intertwined Biogenesis of Photosystems I and II: Born Together to Work Together
Funded by the Grant Agency of the Czech Republic (EXPRO project no. 19-29225X), 2019-2023.
Principal investigator: Prof. Dr. Josef Komenda
Co-investigators: Asoc.Prof. Roman Sobotka, Ph.D. and Radek Kaňa, Ph.D.
Photosystem I and II (PSI a PSII, resp.) are complex enzymes embedded in the thylakoid membrane of cyanobacteria, algae and plants, serving as the key molecular machines of oxygenic photosynthesis (Barber, 2006; Jordan et al., 2001; Umena et al.,2011). The way photosystems are built in the cell is still poorly understood. In a current model, PSII is formed by a combination of modules consisting of a large chlorophyll-protein associated with small subunits and pigments under guidance of specific auxiliary (assembly) proteins (e.g. Komenda, Sobotka and Nixon, 2012). However, the steps of PSI biogenesis have not been described yet. Our recent data indicate that biogenesis of both photosystems is tightly intertwined and performed by common cellular machinery integrating chlorophyll biosynthesis (Dobáková et al., 2009; Bečková et al., 2017; Strašková et al. 2018; Kiss et al., 2019; Knoppová et al., 2022).
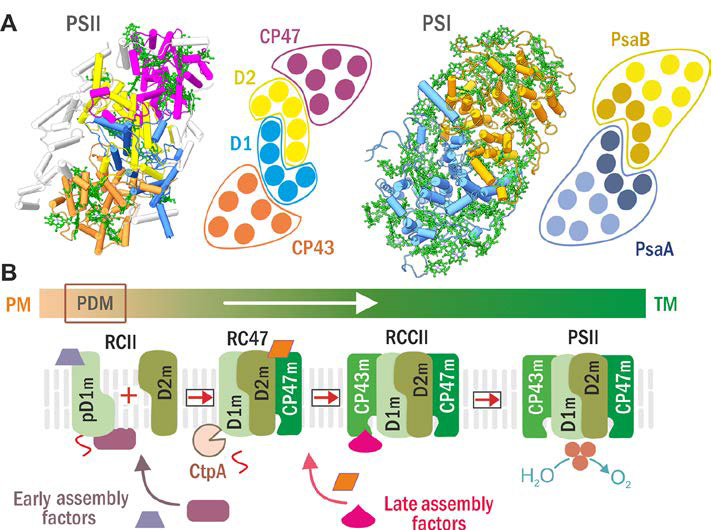
Structures of PSII and PSI (A) and scheme of the current model for PSII assembly and its spatial arrangement in cyanobacterial cells (B). (A) PSII (left) and PSI (right) structure and schematic helical arrangement of their main chlorophyll (Chl)-proteins (D1 in blue, D2 in yellow, CP47 in violet, CP43 in brown; PsaA in blue with antenna helixes in light blue, and PsaB in ochre with antenna helixes in light ochre). (B) The PSII assembly goes on in a stepwise manner starting with the synthesis and insertion of precursor D1 module (pD1m) in so called PratA-defined membranes (PDM; see Schottkowski et al, 2009) close to plasma membrane (PM) and continues with sequential attachment of D2, CP47 and CP43 modules in thylakoid membranes (TM). Formation of RCII complex is accompanied by CtpA-catalyzed pD1 maturation and facilitated by early assembly factors while the later steps by late assembly ones.
As an experimantal organism we employ the cyanobacterium Synechocystis PCC 6803 and a number of its mutants. We use a variety of physiological, biophysical and biochemical methods including purification and 2D analysis of protein complexes combined with protein and chlorophyll radiolabeling, mass spectrometry and confocal and cryo-electron microscopy.
The aim of the project is to verify the hypothesis about intertwined biogenesis of the photosystems and elaborate its model. We also aim to elucidate early phases of PSI biogenesis, including the role of PSI-specific auxiliary factors a determine structure and location of the PSI/PSII biogenesis site.
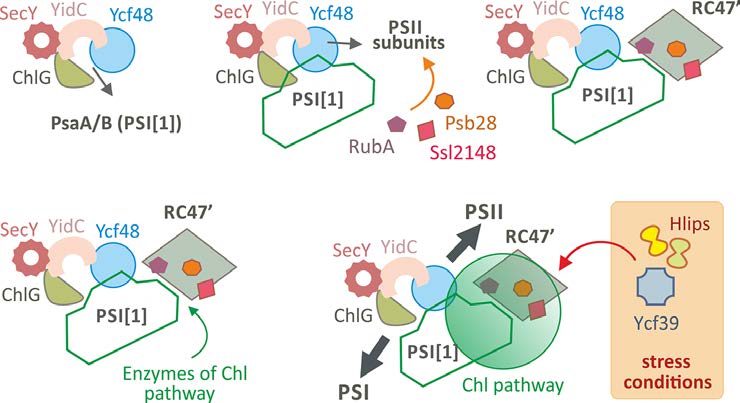
Working model of the biogenesis of Chl-binding components of PSI and PSII in cyanobacteria. Chl-synthase (ChlG) and lumenal Ycf48 proteins are physically attached to SecY translocon via YidC insertase [10] and these proteins form minimal machinery for the synthesis of Chl-proteins. During recovery from a long period of Chl bleaching (e.g. from nitrogen limited conditions) the monomeric PSI (PSI[1]) is preferentially synthesized by utilizing mostly ‘recycled’ Chl molecules and remains attached to ChlG/Ycf48 to serve as a scaffold and photoprotective unit. PSII subunits made later are pre-assembled into RC47 complex and associate with a set of auxiliary factors (Psb28, RubA, Ssl2148 and others). This structural arrangement separates the RC47’ complex from the PSII assembly pathway to work as another structural/functional component of the biogenesis centre. Terminal enzymes of Chl biosynthesis associate with the stromal side of PSI-RC47’ supercomplex, and de novo made Chl precursors are channeled to ChlG to maintain high rate of PSI and PSII Chl-protein synthesis. During stress conditions the biogenetic centre is further equipped with High-light inducible proteins (Hlips) and Ycf39 protein to facilitate Chl re-utilization (Knoppová et al., 2014) and to photoprotect PSII Chl-proteins (Staleva et al., 2015).